Data objects
Or: How do I see the trajectory I generated?
OpenPathSampling stores everything it generates in a single file. This
includes the data generated by the simulation, as well as the objects
describing the simulation itself (enabling easy restarts). When we refer to
storage, we mean the storage subsystem, which deals with how these
things are written to a file. Most users are probably more interested in our
data objects, which are needed for performing custom analysis. The data
objects are described here.
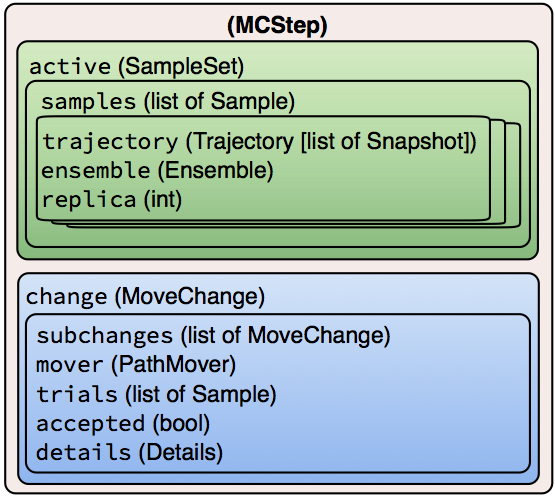
Hierarchical data structure of the MCStep data object. The attribute names are shown in fixed-width font, and the type is provided in parentheses.
The data structures used by OPS allow one to replay the entire simulation,
and this is generally the way we suggest performing analysis: loop over the
steps, in order, and extract the necessary information. The MCStep
object contains information about both the state of the simulation (the
trajectories being sampled) and details on the steps taken during the
simulation. We thing of these as “what is sampled” and “how sampling
happens,” respectively.
The MCStep has two important attributes: .active, which
describes the current (active) state of the simulation at the end of the
given step, and .change, which describes the process that occurred
during this step. These will be discussed in detail below.
Note
Despite the name, MCStep is not only used for Monte Carlo. The
same object is also used for other PathSimulator types, such as
CommittorSimulation. Other simulation types still generate
multiple trajectories, and so the split of “what was the state after this
step” and “how was this step performed” still applies.
Objects describing what is sampled
Snapshots, sometimes called “frames” or “time slices” are at the core of any simulation technique. They describe the state of the physical system at a point in time, and in molecular dynamics, typically consist of coordinates, velocities, and periodic cell vectors. The internal structure of a snapshot is discussed below.A
Trajectory, also called a “path,” is essentially a list ofSnapshotsin temporal order. In addition, it provides several convenience methods, for example, to identify whichSnapshotsare shared by two trajectories.The
Sampleobject is a data structure that links aTrajectorywith theEnsembleobject from which it was sampled, and an integer replica ID. TheSampleis needed because methods such as TIS, and especially RETIS, sample multiple ensembles simultaneously. Correct analysis requires knowing the ensemble from which theTrajectorywas sampled. The replica ID ensures that we can track changes to a given trajectory over time (even if it changes which ensemble it is associated with, e.g., due to replica exchange).Since methods like TIS have several active
Samplesduring a path simulation step, OPS collects them into oneSampleSet. TheSampleSetcontains a list ofSamples, and also has convenience methods to access a sample either by replica ID or by ensemble, using the same syntax as a Pythondict.
Objects describing how sampling happens
The
MoveChangecontains a record of what happened during the simulation step. Because the simulation move itself generally consists of several nested decisions (type of move, which ensemble to sample, etc.), theMoveChangeobject can contain subchanges, which record this entire sequence of decisions. In addition, it includes a pointer to itsPathMover, a list of the trialSamplesgenerated during the step, and a boolean as to whether the trial move was accepted.The
MoveChangealso contains aDetailsobject, which is essentially a dictionary to store additional metadata about a move. This metadata will vary depending on the type of move. For example, with a shooting move, it would include the shooting point. In principle, all the additional information that might be of interest for analysis should be stored in theDetails.
Getting details for the move of interest
The change attribute of an MCStep covers the entire move,
including all the structural elements involved in making the decision. As
such, its details are very general, and not the details (such as shooting
point) that you are probably most interested in.
You can walk through the structural elements using the .subchanges
attribute of a MoveChange, but in order to skip to the details
that you are most likely to be interested in, one MoveChange is
designated “canonical.” For one-way shooting, the change from either the
forward or backward shot is canonical. The change from the replica exchange
mover or path reversal mover or minus mover is canonical. The canonical
change is always within the nested subchanges of the
MoveChange, but can be accessed directly with change.canonical.
Note that this returns a MoveChange; to get the associated
PathMover, use change.canonical.mover. The
change.canonical.details dictionary is where you can find the details of
what happened during this move.
Getting coordinates (etc.) from snapshots
Of course, each Snapshot is a record consisting of several fields, or as
there are referred to in OPS, “features.” Because OPS is independent of the
underlying engine (indeed, the engine need not represent molecular dynamics
at all), these features are engine-dependent. However, we recommend that
particle-based simulation engines use consistent feature names in order to
facilitate integration with tools in OPS and to simplify communication
between engines. These are the features we include for all particle-based
engines in OPS:
coordinates: Positions of the particles with units attached (for engines that have explicit units, such as OpenMM). List of list: the outer list loops over the atoms, while the inner list loops over spatial dimension (typically 3).xyz: Positions of the particles without units attached. Same shape ascoordinates.velocities: Velocities of the particles with units attached (for engines that have explicit units). Same shape ascoordinates.masses: The masses of the system. Units (whether implicit or explicit) should be of actual mass, not mass/mole (as is often done in cases where energies are reported per mole). This may be used to calculate kinetic energy. Shape is the length of abox_vectors: Box vectors for a periodic system, orNoneif system is not periodic. This is usually a 3x3 matrix. OPS uses the same format as MDTraj.engine: The engine instance that created this snapshot. Useful for checking provenance of data.
Note that implementation of these may by such that a single instance is used
by all snapshots. For example, all snapshots generated by a given engine
may share the same list of masses (in order to prevent redundant
storage). However, these are still accessible from the snapshot itself.
Engines with specific needs may include other features. For example, wavefunction information might be included for an engine based on ab initio dynamics. For other features, see the documentation for the specific OPS engine wrapper.
For OPS engines that support it (including the OpenMM engine), trajectories
can be easily converted to MDTraj trajectories with
mdtraj_trajectory = trajectory.to_mdtraj(). From there, one can use all
analysis tools in MDTraj, as well is its ability to write trajectories to
many file formats for input to other analysis programs. In addition, you can
use MDTraj as a gateway to other libraries: for example, its integration
with nglview can be used for
molecular structure visualization.